
|

|
|
| SVM RNAi 2.0: Design siRNA Order Home | |||
|
Incorporated recent advances. Stand-alone SVM RNAi 3.6 is now available. 1.Rational siRNA design by rules and by SVM learning. 2.Designs oligos for a variety of tech platforms. 3.Eliminates unintended targets. 4.Avoids SNP targets. 5.Generates scrambled controls. Click here for more info. SVM RNAi is a learning program. (SVM stands for Support Vector Machine, one of the best statistical learning methods.) It learns from past successes and failures, and builds predictive models for better siRNA design. For more info about SVM and its applications in biology, please visit SVM Classifier and SeqClassifier. |
|
SVM Online runs on Java-enabled browsers PC: Internet Explorer 5.5 or higher and Netscape 4.08 or higher Mac: Internet Explorer 5.0, IE5.1.4 or higher (IE5.1 will not work because of a bug.) |
Need to raise a better antibody? Please use online Abie Pro 3.0 for peptide antibody design. |
|
||||||
|
||||||
| Example V: siRNA/RNAi Design | ||||||
|
Test Drive SVM RNAi 2.0 Stand-alone SVM RNAi 3.6 is now available. Trained with 250 published positive or negative siRNAs, SVM RNAi 3.6 could reduce failure rate by as much as 50%. Click here for more info. 1. Copy (Ctrl-c or Apple-c) and paste (Ctrl-v or Apple-v) the following (or your own) sequence into the input window (top-left) of the SVM RNAi 2.0. Numbers and blanks will be ignored. Note the input sequences should be DNA sequences.
1 tccagcacca aagcggccgt tctcggattc cggagcgttc tggagccccg agagacgccc
61 cggggttcta gaagctcccc ggcggcgccc agtcccggct tcattcgggc gtccctccga
121 aacccactcg ggtgcacggg tcgtcggcga gccgcgaccg ggtcctggcg cgcaccatga
181 tcgtggcgga ctccgagtgc cgcgcagagc tcaaggacta cctgcggttc gccccgggcg
241 gcgtcggcga ctcgggcccc ggagaggagc agagggagag ccgggctcgg cgaggccctc
301 gagggcccag cgccttcatc cccgtggagg aggtccttcg ggagggggct gagagcctcg
361 agcagcacct ggggctggag gcactgatgt cctctgggcg agtagacaac ctggcagtgg
421 tgatgggcct gcaccctgac tactttacca gcttctggcg cctgcactac ctgctgctgc
481 acacggatgg tcccttggcc agctcctggc gccactacat tgccatcatg gctgccgccc
541 gccatcagtg ttcttacctg gtaggctccc acatggccga gtttctgcag actggtggtg
601 accctgagtg gctgctgggc ctccaccggg cccccgagaa gctgcgcaaa ctcagcgaga
661 tcaacaagtt gctggcgcat cggccatggc tcatcaccaa ggaacacatc caggccttgc
721 tgaagaccgg cgagcacact tggtccctgg ccgagctcat tcaggctctg gtcctgctca
781 cccactgcca ctcgctctcc tccttcgtgt ttggctgtgg catcctccct gagggggatg
841 cagatggcag ccctgccccc caggcaccta caccccctag tgaacagagc agccccccaa
901 gcagggaccc gttgaacaac tctgggggct ttgagtctgc ccgcgacgtg gaggcgctga
961 tggagcgcat gcagcagctg caggagagcc tgctgcggga tgaggggacg tcccaggagg
1021 agatggagag ccgctttgag ctggagaagt cagagagcct gctggtgacc ccctcagctg
2. Click on the Predict button to predict. Note the program will predict the core 21 base and sense-strand nucleotide sequences.3. The longest ORF (open reading frame, red bar) and predicted siRNA sequences (green bars) are diagramed on the top-left. Click on a green bar to show the corresponding sequences. 4. Click on the Clear button before running for a different input sequence. 5. Don't forget to run BLAST to check whether a selected siRNA sequence will target uniquely the gene of interest. Or use the stand-alone version which will automatically eliminate targets that are not unique.
How does SVM work? 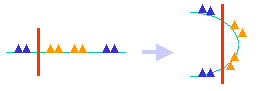 Mathematically the space transformation is achieved by using the kernel functions, which defines distance measures in the new space. In addition, SVM uses only data points close to class boundaries for prediction, thus may be less sensitive to noises in the data. To use SVM, one must first train the SVM to obtain a model. Different model/kernel combinations should be tried to achieve the lowest error rate as determined by the validation. The online version does not allow you to change kernel parameters. You may obtain the stand-alone version if you would like to further improve your model
Please help to improve SVM RNAi You can help us to improve the performance of SVM RNAi. We need more RNAi sequences to train the model. Especially we need sequences that failed in RNA interference experiments. Those sequences will train the SVM to recognize sequences that satisfies rules but nevertheless fail because the rules are not perfect. (Please don't send us your mismatch negative controls.) Please share with us your positive and negative experiences. Your help will be greatly appreciated not only by us but also by future users who will benefit most from your generosity. | ||||||
| Frequently Asked Questions | ||||||
|
How does SVM RNAi increase success rate?
The problem we have is to find a few good siRNA targets. The solution to this problem is simple in principle. All we need to do is to reject more bad siRNA target than good siRNA target, then good targets will be enriched in the remaining pool. Suppose we have 10 good targets among 40 bad targets, if we reject 80% good targets and 95% bad targets, there will be only 2 good targets and 2 bad targets left. We have increased our chance of success by 2.5 folds. To achieve this, we have optimized our SVM machine to reject as many bad siRNA targets as possible even though we may reject many of the good targets. Enrichment in good targets is ensured because bad targets are 1.3-2.5 times more likely being rejected compared to good targets. See Figure 1 for details. Does SVM RNAi use the rules such as AARN17YTT? This is frequently a source of confusion. SVM RNAi is a learning program. It starts with no rules at all. By learning from training data, it comes up with a decision function for testing whether a particular sequence is a good siRNA candidate. The decision function is usually complex and may or may not be interpreted as simple rules. On the other hand, since the majority of the good siRNAs in the training data do satisfy the Tuschlís rules (AARN17Y), a large fraction of targets predicted by SVM RNAi do satisfy Tuschlís rules. However it is not equivalent to say SVM RNAi uses Tuschlís rules. Many sequences satisfying the Tuschlís rules are ruled out by SVM RNAi. Apparently, there are additional restrictions from SVM RNAi. What is the success rate of SVM RNAi? Before talking about success rate, we must define success. We set a high standard for a good siRNA target: (1) the knockdown must be detected at the protein level; (2) the level of knockdown must be reproducible and significant (good for publication). On the training set SVM RNAi achieved a 10-fold cross validation rate of ~90%. On independent sets, the overall accuracy is 50-70% for SVM RNAi 2 and 60%-80% for SVM RNAi 3.6. The overall rate is the success rate in predicting both positives and negatives. SVM RNAi 3.6 is not optimized for best overall success rate. It is optimized to reduce the false-positive rate --- the rate that matters most to researchers. SVM RNAi 3.6 can reduce false-positive rate by as much as 50%. If I choose 100 targets use SVM RNAi 3.6, how many will be good targets? Unfortunately the success rate can't be computed directly from the SVM model. The best estimate we can give is the fail rate: SVM RNAi fail rate ~ 50% * (Tuschl's rule fail rate). Have you trained and tested your model using experimental data that measured mRNA level by real-time RT-PCR? No, we excluded all data that didn't show a knockdown at the protein level. A small decrease in mRNA level may not lead to a significant decrease in protein level since the translation machinery may compensate for the decrease in mRNA. If a siRNA also suppresses translation, then the change in the protein level will be greater. Furthermore, antisense RNA may also cause a decrease in the mRNA level as measured by real-time RT-PCR (Vickers et al, J Biol Chem. 278(9):7108-18). What do you mean SVM RNAi 3.6 can reduce false-positive rate by as much as 50%? If you have designed five siRNAs using the rules most people have used and only one showed specific siRNA activity, your false-positive rate is 4/5=80%. Reducing the false-positive rate by 50% means that the new false-positive rate will be 80% * 50% = 40%, i.e., only 2 (5 * 40%) siRNAs will not have specific activities if five targets are selected by SVM RNAi 3.6. If I reject all siRNA candidates, my false-positive rate will be 0. SMV RNAi does not randomly reject siRNA targets. It is tested to ensure that the probability of rejecting a good siRNA target is less than the probability of rejecting a bad siRNA target. As a result the remaining pool of siRNAs are enriched in good siRNA targets. See Figure 1 for detailed statistics. Why do I need the stand-alone version? The stand-alone SVM RNAi is version 3.6 while the online one is version 2.0. Version 3.6 is trained and tested with a larger set of published siRNA sequences compared to version 2.0 (~300 verses ~100 siRNA sequences). The stand-alone version allows users to eliminate siRNAs with multiple targets. Blast may also be used for this step. Blast is time consuming and results may not be clear especially when close family members exist. It is easy to overlook Blast results. Why do you eliminate siRNAs with only 15-base match to more than one gene? Although a single mismatch may abrogate silencing, recent findings suggest that shorter matches may translationally repress untargeted genes (Semizarov et. al., PNAS 100(11), 6347-6352; Doench et. al., Genes Dev. 17(8):991-1008; Jackson et. al., Nature Biotech. 21(6): 635-637). Furthermore 15-base match to untargeted genes may nevertheless decrease effective siRNA concentration. For this reason any siRNA duplex (in two directions) with a potential of binding to more than one gene will be eliminated. What is the user experience? User feedbacks so far are in general positive. Performances of SVM 3.6 on data provided by users are better or as good as on published data. Does SVM RNAi work for hairpin siRNA? There is no strong reason to suspect hairpin siRNA will have a different set of rules for target selection. The SVM model is not trained separately for hairpin siRNA. How do I get SVM RNAi 3.6?
| ||||||
| More SVM Examples | ||||||
| Please visit SVM Classifier and SVM SeqClassifier for more examples. | ||||||
THIS SOFTWARE IS PROVIDED BY CHANG BIOSCIENCE ``AS IS'' AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL CHANG BIOSCIENCE OR ITS SPONSORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
